Simone Bendazzoli

About me
I began my PhD in October 2020, focusing on medical image processing using deep learning — a field that has rapidly evolved but still faces challenges in real-world clinical adoption. Over the years, working at the intersection of AI research and medical practice has made me reflect deeply on how difficult it remains to integrate the models we develop into clinical workflows in a transparent, reproducible, and scalable way.
This realization led me to design and build MAIA, an open-source Medical AI platform that promotes collaboration and reproducibility in medical imaging research. MAIA integrates Kubernetes, MONAI, and the MONAI Bundle format to simplify deploying, configuring, and updating AI models in distributed research environments. It integrates essential tools such as 3D Slicer, OHIF Viewer, Orthanc, XNAT, and MLFlow, forming a cohesive ecosystem for annotation, model training, active learning, and refinement.
Through this work, I aim to bridge the gap between AI innovation and clinical implementation by providing researchers and clinicians with the infrastructure they need to collaborate effectively.
As I approach the defense of my PhD thesis on November 27, 2025, my focus remains on advancing open, modular, and transparent medical AI systems that can empower the community and accelerate the safe translation of research into practice.
When I’m not coding or experimenting with new infrastructure designs, I’m usually out running — my favorite way to clear my mind, stay focused, and find inspiration for the next challenge. Follow my running activities on Strava:
Resume
PhD Dissertation
I will be defending my PhD thesis at KTH on Thursday, November 27, 2025. My dissertation is entitled “Design and Integration of AI Solutions in Oncology and Healthcare Infrastructures: Bridging the Gap Between AI Innovation and Clinical Practice”. During my defense, I will present the results of five years of research, covering the creation of MAIA, the development of the MONet Bundle, and advancements in federated and active learning.
You can access the public thesis via these links in KTH DiVA and KI Open Archive:
Simone Bendazzoli
KTH Royal Institute of Technology, Karolinska University Hospital & Karolinska Institutet, Stockholm, Sweden
November 27, 2025, 09:00 CET, T2 (Jacobssonsalen), Hälsovägen 11C, Huddinge, Sweden
Research
MAIA: A Collaborative Medical AI Platform for Integrated Healthcare Innovation
Simone Bendazzoli, Sanna Persson, Mehdi Astaraki, Sebastian Pettersson, Vitali Grozman, Rodrigo Moreno
KTH Royal Institute of Technology, Karolinska University Hospital & Karolinska Institutet, Stockholm, Sweden
The integration of Artificial Intelligence (AI) into clinical workflows requires robust platforms that bridge the gap between technical innovation and practical healthcare applications. MAIA (Medical Artificial Intelligence Assistant) is an open-source platform designed to facilitate interdisciplinary collaboration among clinicians, researchers, and AI developers.
Built on Kubernetes, MAIA provides a modular, scalable environment with integrated tools for:
- Data management
- Model development
- Annotation and active learning
- Deployment and clinical feedback
Key features include project isolation, CI/CD automation, and integration with high-computing infrastructures and real-world clinical workflows. MAIA supports deployments in both academic and clinical environments, enabling reproducible, transparent, and user-centered AI development.
MAIA has been applied in multiple medical imaging projects at KTH Royal Institute of Technology and Karolinska University Hospital, demonstrating its ability to accelerate the translation of AI research into impactful clinical solutions.
- Article: https://www.nature.com/articles/s44387-025-00042-6
- Preprint: https://arxiv.org/abs/2507.19489
- More details: MAIA page
GitHub Code: MAIA
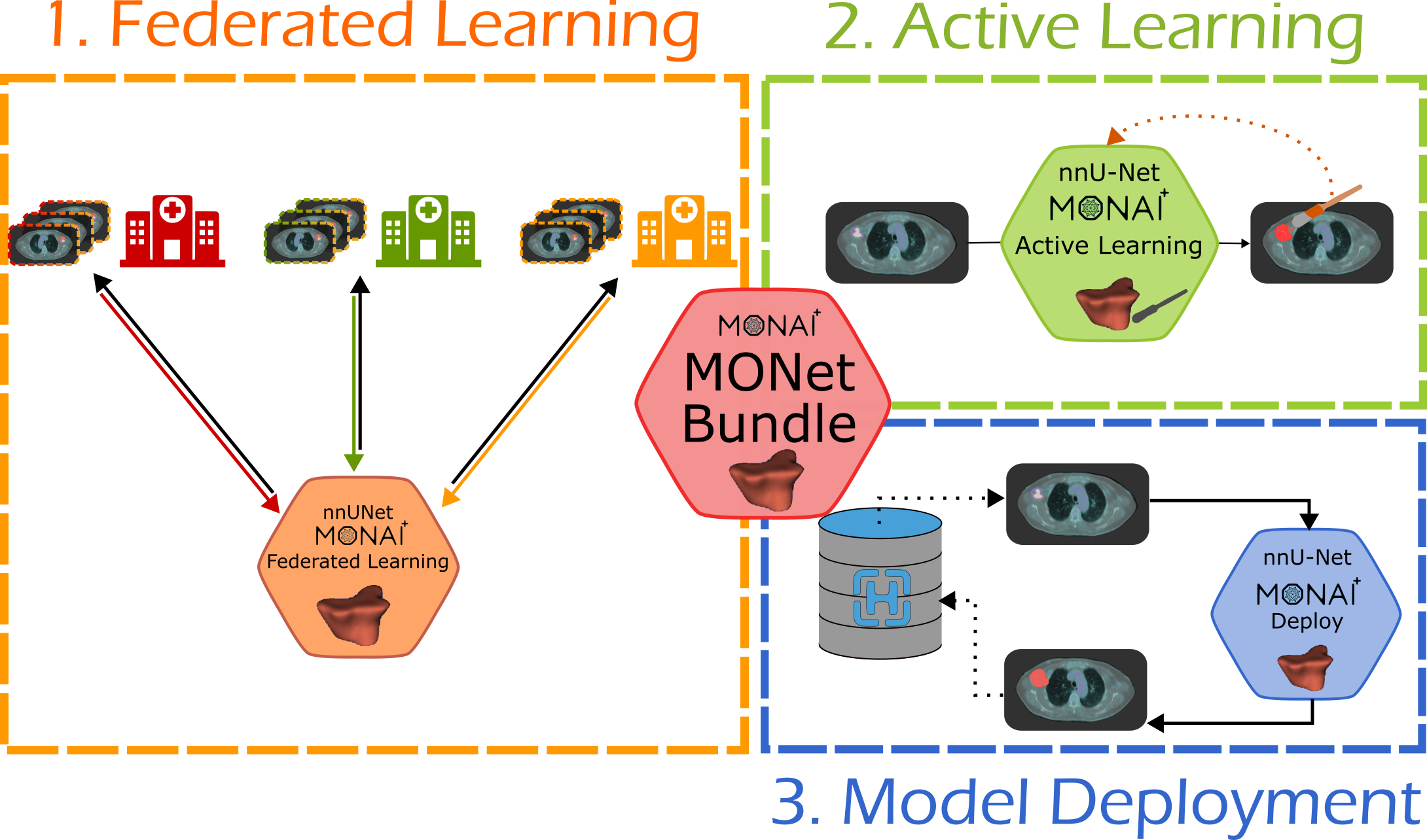
MONet Bundle: Integrating nnU-Net within the MONAI Ecosystem
MONet is a nnU-Net-based MONAI Bundle designed to bridge the gap between state-of-the-art medical image segmentation research and real-world clinical deployment. While nnU-Net is widely recognized for its robustness, versatility, and self-configuring capabilities across diverse medical imaging tasks, its native implementation faces challenges in clinical settings, such as limited portability, adaptability, and integration with existing hospital workflows.
The MONAI ecosystem provides a standardized framework for training, deployment, and annotation of medical AI models, including tools like MONAI Deploy for clinical integration and MONAI Label for AI-assisted image annotation. Starting from MONAI v1.2.0, nnU-Net has been partially integrated via nnUNetV2Runner, enabling data preparation, preprocessing, training, and cross-validation within MONAI pipelines. However, deployment and packaging for diverse clinical applications remained limited.
MONet Bundle extends nnUNetV2Runner to:
- Package nnU-Net models into versatile bundles for multiple tasks.
- Enable federated learning and multi-center collaboration.
- Integrate with clinical workflows, improving portability and adaptability.
This ongoing effort contributes to both nnU-Net and MONAI repositories (MONAI Core, MONAI Deploy, and MONAI Label), providing a scalable and reproducible framework that facilitates the transition of segmentation models from research to real-world clinical use.
Applications and Achievements:
- Competed in BraTS 2025 Challenge Task 07: GoAT – Generalizability Across Tumors, where MONet Bundle achieved 1st prize, demonstrating robust performance across adult glioma, brain metastasis, meningioma, pediatric glioma, and sub-Saharan African cohorts.
-
Used in a federated learning approach for whole-body PET-CT lymphoma segmentation, presented at the DeCaF 2025 Workshop at MICCAI. Paper link
Code: https://github.com/minnelab/MONet-Bundle
Documentation: https://minnelab.github.io/maia/
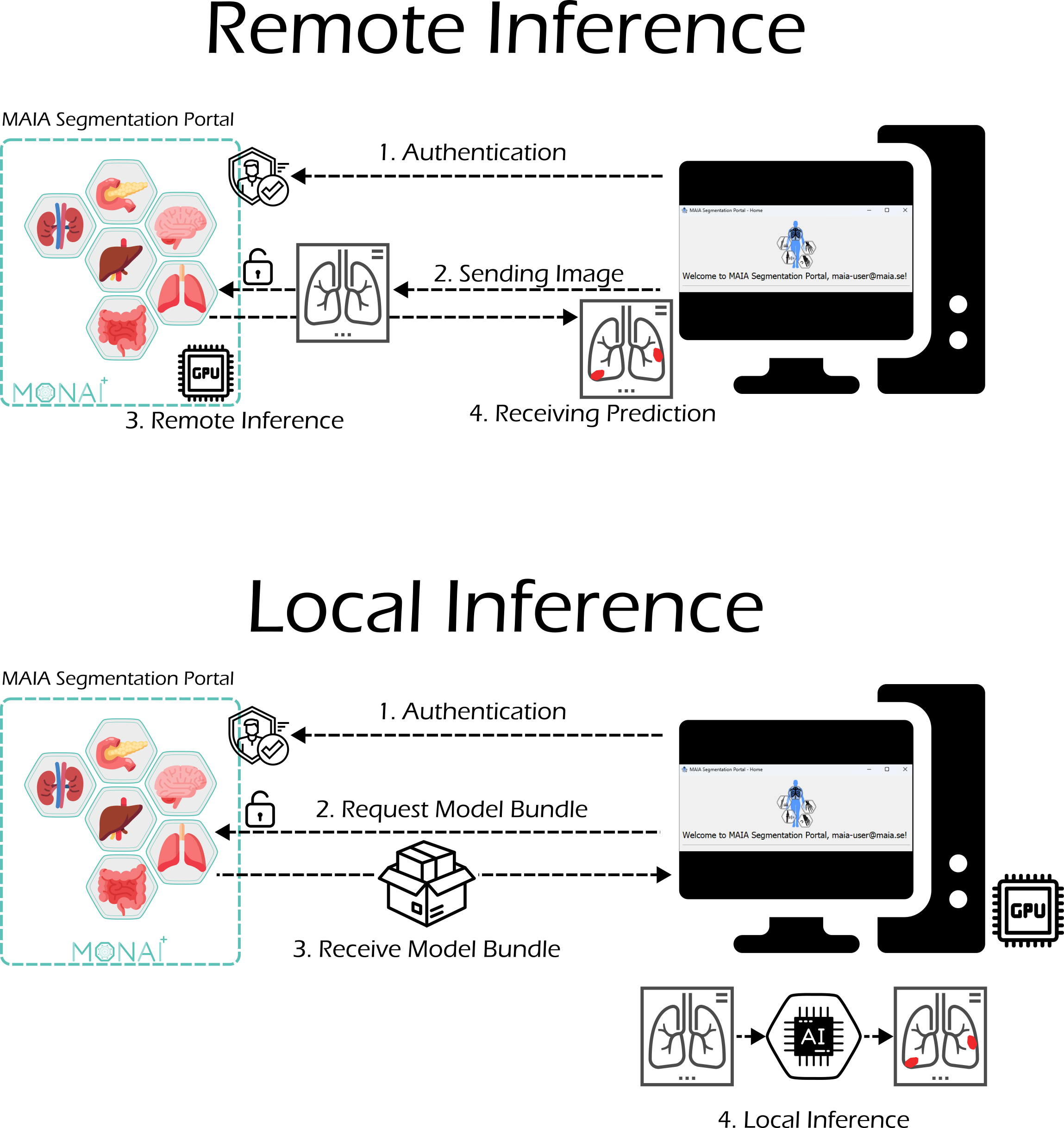
MAIA Segmentation Portal
The MAIA Segmentation Portal is a user-friendly platform hosted within MAIA, allowing users to explore and interact with available medical AI models. Users can upload medical images and receive predictions almost instantly, or download the models to run locally if preferred.
The portal leverages models in the MONet Bundle format, which are compatible with MONAI Deploy and MONAI Label, making it easy to integrate AI into clinical workflows. Each model is deployed as a standalone application within MAIA, and the portal is built on a KubeFlow-based infrastructure, allowing smooth interaction and inference directly from the portal interface.
The MAIA Segmentation Portal also provides guidance for users who want to train their own models using their annotated data, promoting reproducibility and collaboration in medical AI research.
Documentation: MAIA Segmentation Portal Documentation
Lung Vessel Connectivity Map as Anatomical Prior Knowledge for Deep Learning–Based Lung Lobe Segmentation
Simone Bendazzoli, Emelie Bäcklin, Örjan Smedby, Birgitta Janerot-Sjöberg, Bryan Connolly, Chunliang Wang
KTH Royal Institute of Technology, Karolinska Institutet, Karolinska University Hospital
J. Med. Imaging, 2024 — PMID: 38988990 | DOI: 10.1117/1.JMI.11.4.044001
This study explores how anatomical priors can enhance deep learning segmentation of lung lobes from chest CT. We introduced a Lung Vessel Connectivity (LVC) map, encoding the vascular structure of the lungs, and integrated it into three nnU-Net variants: standard, multitask, and cascade models.
Including the LVC map improved segmentation accuracy, especially at challenging interlobar boundaries and in expiration scans, while also improving robustness in pathological cases such as COVID-19. The results demonstrate that embedding anatomical knowledge into model design can enhance both accuracy and generalization, promoting more clinically reliable segmentation systems
Designing Radio-dynamics Features for pCR Prediction in Breast DCE-MRI
Simone Bendazzoli, Mehdi Astaraki, Yanbo Li, Rodrigo Moreno, Örjan Smedby, Hong Lu, Chunliang Wang
KTH Royal Institute of Technology, Karolinska Institutet, Tianjin Medical University Cancer Hospital
This work introduces a novel family of imaging biomarkers, radio-dynamics features, designed to improve pathological complete response (pCR) prediction in breast cancer patients undergoing neoadjuvant chemotherapy (NAC). These features are derived from dynamic contrast-enhanced MRI (DCE-MRI) and aim to capture the temporal dynamics of tumor enhancement beyond what traditional radiomics can represent.
The study implemented a full machine learning pipeline for feature extraction, selection, and classification, evaluated on 80 subjects using 5-fold cross-validation. Results showed that radio-dynamics features outperformed conventional radiomics, particularly in ensemble models, demonstrating their potential for more accurate and physiologically meaningful prediction of treatment response.
The source code and pipeline are openly available at: https://github.com/minnelab/Hive_ML